In this update, the Mapping Cancer Markers research team explains how a relatively small code change could have a big impact on the project’s ability to analyze markers for various types of sarcoma.
Background
Mapping Cancer Markers aims to identify the markers (sometimes referred to as signatures) associated with various types of cancer. The project is analyzing millions of data points collected from thousands of healthy and cancerous patient tissue samples. So far, these have included tissues with lung cancer, ovarian cancer, and sarcoma.
Expanding our ability to work with data sets
Code changes are coming soon to improve the Mapping Cancer Markers project. These changes will expand the Mapping Cancer Markers application's ability to deal with multi-label data sets and allow it to search for more specific signatures within them.
Figure 1A
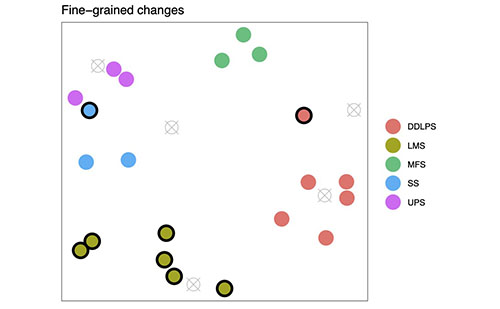
Figure 1A (above): This represents the sarcoma dataset, with seven subtypes of samples. Mapping Cancer Markers can use the multi-labelled dataset as-is and search for multiclass (one-against-all) signatures. (The project software can do this now.)
Before the sarcoma data set, the project analyzed lung and ovarian cancer data sets, both of which have binary labels. Our lung cancer data set labelled samples as either "cancer" or "no cancer." Our ovarian data set labelled samples as short or long survival. The sarcoma data set is multi-label, and labels samples with seven different subtypes of sarcoma (see Figure 1A above).
Figure 1B
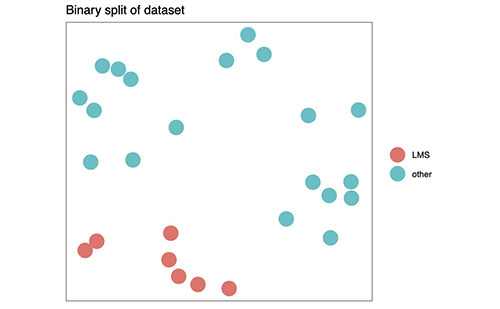
Figure 1B (above) shows how MCM can reduce sarcoma to a binary dataset, by splitting the subtypes into two groups and searching for binary signatures. (The project software can do this now.)
Figure 1C
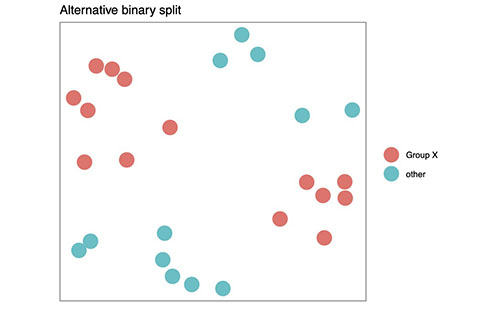
Figure 1C shows an alternative reduction to a binary dataset. (The project software can do this now.)
When a dataset has binary labels, Mapping Cancer Markers will find signatures that can predict that binary label. With a multi-label data set, however, we can direct Mapping Cancer Markers to search for either binary or multiclass signatures (see Figures 1B and 1C above for examples). Presently, the project is searching for both in the sarcoma data set. A multiclass sarcoma signature distinguishes every subtype from every other; given any sarcoma sample, it will diagnose the specific cancer subtype. A binary sarcoma signature distinguishes one group of subtypes from the rest, but does not distinguish specific subtypes. For example, among the seven sarcoma subtypes are two leimyosarcoma (LMS) subtypes, soft-tissue and uterine. Mapping Cancer Markers is presently searching for binary signatures that distinguish LMS from the rest.
Figure 1D
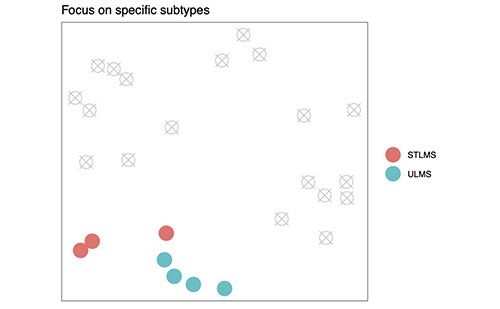
Figure 1D shows how Mapping Cancer Marker's new capabilities will allow a work unit to focus on specific subtypes. Excluded samples are grey and crossed-out. (New code.)
Figure 1E

Figure 1E shows the Mapping Cancer Marker's new capabilities in full, excluding individual samples and changing their labels. Relabeled samples are outlined in black. (New code.)
While planning the project’s transition to sarcoma, we realized our sarcoma experts had scientific questions about sarcoma they wanted to explore that required more flexibility in work unit design than was possible with the existing application. They wanted to explore differences between two or more specific subtypes of sarcoma, and exclude others from analysis (see Figure 1D). (For example, they wanted to explore biomarkers that distinguish LMS subtypes.) Unfortunately, the ability to exclude samples was not built-in to the original Mapping Cancer Markers application.
Working together, the Mapping Cancer Markers team designed a small extension to the application that would add the needed capabilities, provide additional flexibility for future needs, and preserve backwards compatibility, while minimizing total code changes (Figure 1E).
In the recent months, World Community Grid volunteers have processed thousands of work units to beta-test the new code. These changes give us the power to make fine-grained adjustments that tailor the dataset to the precise question each work unit will explore.
Thank you to everyone who is supporting Mapping Cancer Markers.