How does CPU and GPU compare in terms of energy and speed? OPN researchers compare them using two example packages.
AutoDock 4 (AD4) used in Open Pandemics project (OPN1) uses a genetic algorithm as the global search to cover a large parameter search space in conjunction with an energy-based Solis-Wets (SW) local search to find poses with the lowest energy. For each ligand, we specify a certain number of energy evaluations based on the number of atoms and how many rotatable bonds are used. Typically, the number of evaluations grows exponentially with the number of rotations.
AD-GPU used in OPN1-GPU (OPNG) uses AD4's genetic algorithm but, additionally to the SW local search, it also implements the gradient-based Adadelta local search. Adadelta is able to find better poses (lower energy) with less energy evaluations compared to SW, however, this comes at typically a 10x higher computational cost. In order to mitigate some of this additional cost we came up with the AutoStop algorithm that constantly monitors the docking calculations for statistical signs of convergence and, when it is "reasonably sure" a given set of runs is converged, it can stop a given docking earlier.
For a typical mix of ligands, AutoStop saves about 10x of runtime by reducing the overall evaluations, allowing us to get the best of both worlds: accuracy and speed.
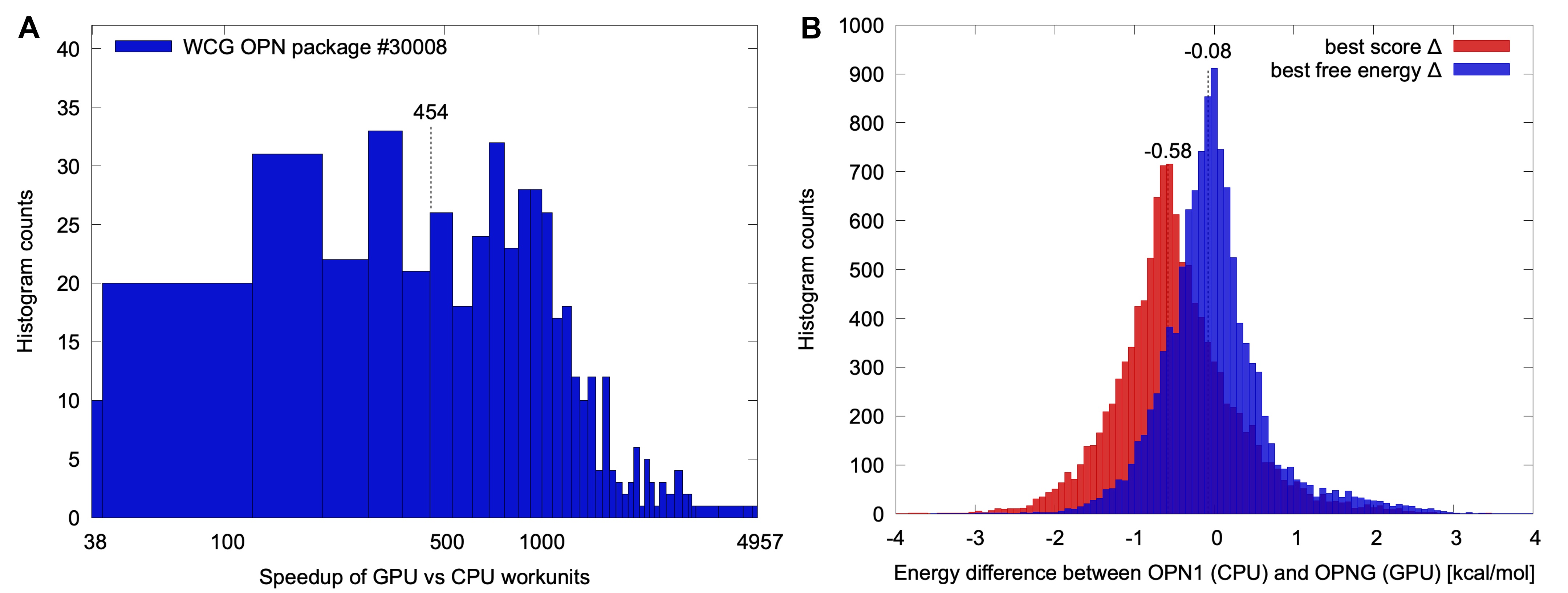
Figure 1 shows a comparison obtained running OPN1 package number 8 with the shown GPUs using the current OPNG approach. The OPN1 runtimes were normalized to a single core of an AMD Ryzen 9 3900x which was one of the fastest CPUs on WCG during this package's run.
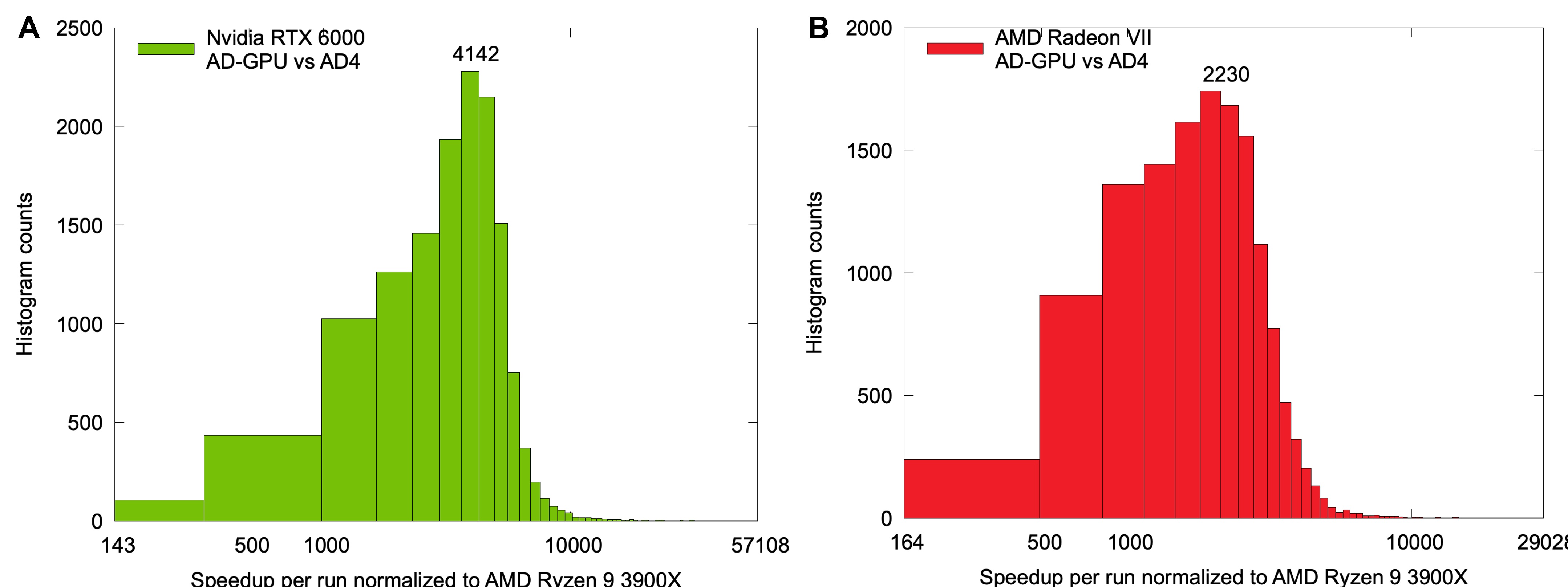
Figure 2 shows the overall speedup with a mix of CPUs and GPUs from WCG during the beta test for OPNG using OPN1 package 30008. Note how AD-GPU gives better energy estimates than AD4 as we use improved algorithms.