The OpenZika team has published a new research paper outlining the progress they have made after their partnership with WCG
Brief history of the OpenZika project
Dr. Carolina Horta Andrade (Faculdade de Farmácia Universidade Federal de Goiás, LabMo), together with Dr. Sean Ekins (Collaborations Pharmaceuticals, Inc.) and Dr. Alexander Perryman (Rutgers University–New Jersey Medical School) started the OpenZika project in May of 2016 in collaboration with IBM WCG to identify possible inhibitors of the Zika virus. Patients affected by this virus suffer paralysis of their nervous systems and children born to mothers affected by the virus have severe brain development defects. While it was already an enormous threat in Brazil, it had the potential to become a global threat unless a treatment was developed
quickly. WCG’s massive computational power enabled the team to rapidly test millions of compounds in search of possible inhibitors of various Zika virus targets.
By November, the list of 7,600 potential compounds was screened to test their efficacy against the NS3 helicase, a Zika virus protein that allows it to unwind its duplex RNA. The list of molecules was reduced to eight compounds, five of which were subsequently verified to be safe by a study conducted at the University of California, San Diego. In March of 2017, the team was ready to move onto the second stage of the project. The team used a server to create a large library of 30.2 million compounds to be tested against the NS2B-NS3 protease, NS3 helicase, and NS5-polymerase proteins. Running computational screening on this large library was only possible thanks to the combined efforts of the World Community Grid (WCG) volunteers.
The tests on these 30.2 million compounds continued through December of 2018, when the team analyzed an additional compound database supplied by ChemBridge. The list of one million compounds was tested in their effectiveness against NS5 polymerase, NS5 methyltransferase, and the NS3 helicase; ultimately reducing it to 55 compounds of interest. The results were sent to the University of California for evaluation in the virus, as well as to University of Sao Paulo for evaluation with the Zika virus proteins in July, 2019.
Powered by a community of 80,000 volunteers who donated nearly 73 CPU years a day on average to the WCG project, the OpenZika team was able to finalize their modeling and the project was completed in December 2019. The next phase was focused on the experimental validation and prioritization of the selected molecules.
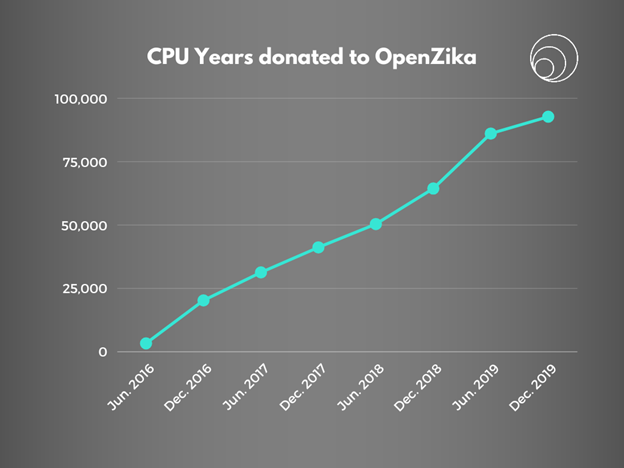
Figure 1: Total CPU years donated to the OpenZika project had been steadily increasing since May 2016 till December 2019.
New developments
In October 2022, the team’s four years of research culminated with a publication in the Journal of Chemical Information and Modeling[1].
The paper highlights the computational process and the validation steps performed during and after WCG analysis. The research team published the results for three important Zika virus proteins: NS3hel, NS2B-NS3pro, and NS5 RdRp. Using WCG, the researchers searched millions of commercially available compounds and identified 61 possible compounds of interest for further screening and optimization.
After the massive docking calculations, the compounds were filtered using machine learning models developed by the LabMol group to identify those that were cytoprotective against Zika Virus infection. Then, compounds that were predicted to pass the blood brain barrier were retained, in order to select those that could counteract the effects the virus has on the central nervous system. Finally, a medicinal chemistry inspection identified and selected compounds with desirable features present in existing drugs.
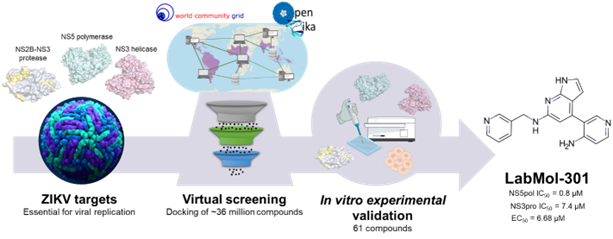
Figure 2: Description of the discovery pipeline; created by Bruna K. P. Sousa from Dr. C. Horta Andrade lab
From almost 404 million results generated by WCG, amounting to almost 93 thousand years of computation, 61 hits were prioritized for testing. Using enzymatic and phenotypic assays, five compounds were ultimately selected as they inhibited the function of or destabilized the three viral proteins of interest, NS2B-NS3 protease, NS3 helicase and NS5-polymerase proteins. Further tests identified 8 compounds as being able to protect the cells from death caused by the virus, while showing low cellular toxicity in liver and kidney-derived cells. The two sets of 5 and 8 molecules overlap for two compounds, named by the authors LabMol-301 and LabMol-212.
Dr. Carolina Horta Andrade: “This work has demonstrated the importance of the integration of computational and experimental approaches, as well as the potential of large-scale collaborative networks to advance drug discovery projects for neglected diseases and emerging viruses, despite the lack of available direct antiviral activity and cytoprotective effect data, that reflects on the assertiveness of the computational predictions.”
The results of this research are exciting, and further optimization could lead to these molecules being tested as antiviral treatments for Zika virus. The researchers are now looking for partners for performing the hit-to-lead optimization with chemical synthesis and further experimental validations.
We thank the volunteers who made these findings possible, and to the OpenZika team for sharing this exciting update and their continued involvement with WCG. If you have any comments or questions, please leave them in this thread for us to answer. Thank you for your support.
WCG team
[1] Melina Mottin, Bruna Katiele de Paula Sousa, Nathalya Cristina de Moraes Roso Mesquita, Ketllyn Irene Zagato de Oliveira, Gabriela Dias Noske, Geraldo Rodrigues Sartori, Aline de Oliveira Albuquerque, Fabio Urbina, Ana C. Puhl, José Teófilo Moreira-Filho, Guilherme E. Souza, Rafael V. C. Guido, Eugene Muratov, Bruno Junior Neves, João Hermínio Martins da Silva, Alex E. Clark, Jair L. Siqueira-Neto, Alexander L. Perryman, Glaucius Oliva, Sean Ekins, and Carolina Horta Andrade. Discovery of New Zika Protease and Polymerase Inhibitors through the Open Science Collaboration Project OpenZika. Journal of Chemical Information and Modeling, 62(24), 6825-6843, 2022. DOI: 10.1021/acs.jcim.2c00596
https://pubs.acs.org/doi/full/10.1021/acs.jcim.2c00596