The software that powers OpenPandemics - COVID-19 has been adapted to use GPU power. This will not only help the researchers to screen more molecules, but also could help them study molecules that are more complex.
For several months, the OpenPandemics - COVID-19 researchers in the Forli Lab at Scripps Research and World Community Grid's tech team have been working behind the scenes to adapt AutoDock 4, the software that powers the project, for use on devices with graphics processing units (GPUs).
During beta testing, we found that work units running on volunteers' GPUs finished an average of 500 times faster (GPU elapsed time vs CPU time) than the same amount of work when it was run using the CPU version of AutoDock.
What is a GPU?
A graphics processing unit (GPU) is another name for the graphics card inside a computer originally used to display text and images on a monitor. As GPUs evolved they gained vast amounts of parallel processing power. Initially, this was a by-product of accelerating specialized graphics related calculations but eventually manufacturers extended these capabilities to allow general purpose computations. As a consequence, modern GPUs can perform certain types of computations significantly faster than the central processing units (CPUs) that orchestrate all work in a computer. In order to utilize a GPU for scientific calculations, however, applications have to be modified to be able to utilize high levels of parallelism to leverage the GPUs’ computational capabilities efficiently.
If a volunteer runs OpenPandemics - COVID-19 on a device that has a suitable GPU, and adjusts their settings accordingly, that device can complete work units much more quickly than a device using just a CPU.
Why is it important to increase the speed of this project?
AutoDock-GPU (AD-GPU) could increase the project's chances of finding a molecule with anti-viral properties even further.
Compared to AutoDock 4 (the current version of the software that is used for OpenPandemics - COVID-19), AD-GPU is much faster which will provide a nice boost to the already amazing rate of docked results.
Furthermore, AD-GPU has an improved search algorithm that exhibits a greater probability of finding strong interactions between the molecules and viral proteins, and is well suited to dock larger or more complex molecules. This means that the researchers can use AD-GPU to not only screen more molecules, but also to enable the search of more complex molecules.
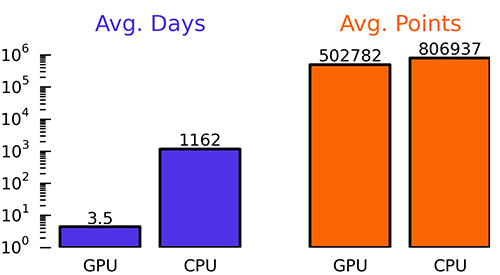
Comparison of OpenPandemics GPU to CPU for the originally CPU-targeted batches 30010-30019 run during beta testing. Average Days to complete one entire batch (blue bars) are shown with Average Points per batch (orange bars). Overall average speedup was 334 times (maximum observed was 516x). The overall average points per batch highlight a 1.6x increase in the algorithmic efficiency of the GPU. This increased efficiency will be leveraged in GPU-targeted packages and lead to much higher points per GPU-batch.
More molecules, more complex molecules, and in less time...this is all crucial to help find potential treatments for a virus that is not only still spreading in most of the world but is also continuing to mutate.
So are devices without GPUs still important for this project?
Yes! Currently, only about 20 percent of World Community Grid power comes from devices with GPU, so participation from every willing CPU-enabled computer, Android device, and Raspberry Pi remains crucial.
Thank you to everyone who is supporting OpenPandemics - COVID-19!
To learn more about GPU power for this project--including how to make your GPU-enabled device can participate--visit our GPU FAQs.