In this extensive update, the FightAIDS@Home team leader, Prof. Art Olson, recaps nearly a decade of progress in the fight against AIDS: new computational methods, new understanding of key HIV proteins, and huge volumes of computational results that have only begun to be explored. Though Phase 1 is winding down, Phase 2 of this enormous project will continue to advance vital research into the world's deadliest virus.
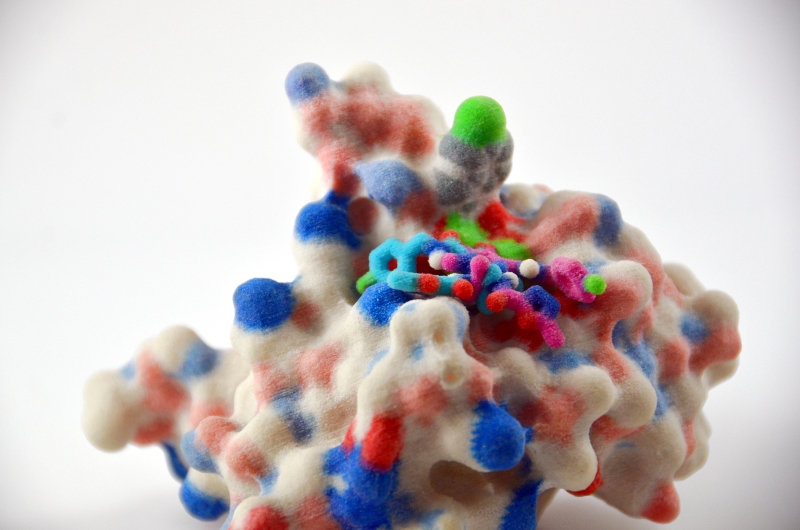
The FightAIDS@Home (FAAH) project joined World Community Grid almost 10 years ago, and is its longest continuously running project. Phase 1 is currently winding down, so it's a good moment to give World Community Grid volunteers a summary of who we are, what our goals have been, what computational approaches have been enabled by the resources you have provided, and what we've accomplished to this point. We're continually aware that this research is only possible because of your incredible generosity, and we hope you will all stay involved in Phase 2 of our work.
Who is behind FightAIDS@Home?
FAAH, which uses computational studies to help advance the search for new treatments against HIV/AIDS, began on Entropia (a now-defunct distributed computing platform) in 2000 as a collaboration between Scott Kurowski, Tim Cusac, Dr. Garrett Morris, and myself. On November 21, 2005, FAAH joined World Community Grid. During the last decade, the constantly increasing computing power provided by World Community Grid volunteers enabled us to expand the scale of our HIV research by many orders of magnitude. As an added bonus, these tremendous resources also inspired us to create new tools and strategies to harness the potential provided by World Community Grid. These tools are not only increasing the efficiency and accuracy of our own computationally-driven studies against HIV, but also help advance the research that the rest of the scientific community pursues against other diseases.
The FightAIDS@Home project has been run in the Olson Laboratory by a dedicated succession of Postdoctoral Research Associates over the past 10 years. Garrett Morris, William "Lindy" Lindstrom, Alex Perryman, and Daniel Santiago have in turn taken the lead in running our computational experiments on World Community Grid. They have been assisted over the years by several lab members including Michael Pique, Ruth Huey, Stefano Forli, Sargis Dallakyan, Alex Gillet, and Max Chang. As in most academic labs, postdocs stay for a period of 2 to 4 years and then move on to more permanent positions. Daniel Santiago has just left the lab, and we are currently interviewing for a new Postdoctoral Fellow to take his place.
FightAIDS@Home goals
The Human Immunodeficiency Virus (HIV), the virus that causes Acquired Immunodeficiency Syndrome (AIDS), has infected over 70 million people throughout the world since this pandemic began. Currently, over 35 million people are infected with HIV, and there are over 2 million new infections each year. Almost one million people die from AIDS each year, which makes HIV the deadliest virus that afflicts humanity.
The Olson Lab is interested in understanding the molecular mechanisms responsible for the evolution of drug resistance in HIV, and in using that understanding to help develop new therapeutic approaches to treat HIV-infected individuals. I direct an NIH-funded center called the HIVE (HIV Interaction and Viral Evolution) Center, which is composed of 14 individual laboratories in 8 different institutions around the country. The Olson laboratory, as well as that of Prof. Ron Levy at Temple University, is involved in the computational aspects of the goals of the Center. Our approach to the problem of drug resistance has been to simulate the interactions of synthetic organic molecules with viral proteins to predict which ones might bind to and affect the activity of these proteins. To do this, my Lab has developed and use two computational docking programs AutoDock and AutoDock Vina to screen large libraries of chemical compounds. The primary goal in these computations is to discover which compounds can potentially interact with and inhibit the activities of the so called "wild type" and drug resistant mutations of the proteins that have evolved in treated patients. The primary protein targets for this work have been the HIV protease (PR), integrase (IN), and reverse transcriptase (RT). These are the three proteins for which the FDA has approved drugs for the treatment of HIV-infected patients. They are also the three enzymes that the virus produces to carry out the chemical reactions that sustain the viral life cycle. By using the extensive structural data on wild type and mutant proteins, as well as mutational data from treated HIV-infected patients, we explore hypotheses and relationships between molecular structure and the development of drug resistance. The computational resources of World Community Grid have been invaluable in extending our computational approaches in this research effort.
In the initial years of FAAH, experiments generally involved computationally evaluating a few thousand compounds against one pocket of one type of the viral machinery - the active site of HIV protease. The "active site" is the region where the chemical work gets accomplished by an enzyme. HIV protease chops the long, viral, multi-protein polypeptide chain at several different, specific places. By cutting that multi-protein chain into different pieces, it then allows those different, individual proteins to fold up into their normal, mature shapes that are required for them to function. Those folded viral proteins work together to create new HIV particles that escape the infected cell, mature, and then infect new cells, which spreads the infection within the patient and allows the infection to spread to new people. When HIV protease activity is sufficiently disrupted, then those multi-protein viral polypeptide chains are no longer separated in an efficient, well-ordered manner. This causes the infected cell to produce immature, non-functional particles that are not able to infect other cells. Of greatest importance, when HIV protease drugs were combined with HIV reverse transcriptase drugs, the death rate associated with HIV infections drastically decreased. Before HIV protease drugs existed, getting HIV was basically a rapid and horrible death sentence. When HIV protease drugs were combined with other classes of anti-HIV drugs to make the HAART cocktails (Highly-Active Anti-Retroviral Therapy), then many HIV patients were able to live long, productive lives with a reasonable quality of life. However, since HIV keeps evolving into slightly different forms that are able to resist the effects of these drugs (that is, new multi-drug-resistant mutant superbugs keep appearing and spreading), we need to discover new types of drugs and therapeutic interventions that can disable these mutants.
World Community Grid enables unprecedented computational approaches for FAAH
The computational docking problem is a high-dimensional search of possible interactions between two molecules, a chemical compound and a target viral protein. One must typically generate millions of poses of the two interacting molecules, and for each pose compute the energy of interaction between them. This energy evaluation is accomplished by summing up the interaction energies between all of the atoms in the two molecules. When we first developed AutoDock in 1990, a single docking would take 20 minutes or more on the fastest computers of that time. Today, thanks to Moore’s Law, such a calculation may take a few seconds to a minute depending on the complexity of the molecules involved. Thus, the ability to screen many molecules against a specific protein target (i.e. virtual screening) became possible. However, finding a good candidate molecule (a hit) that binds well to a functional site on a protein requires screening a very large number of chemical compounds. The larger the library of compounds, the better the chances that a good hit compound will be in that library.
Having our FAAH project on World Community Grid has enabled us to run computations that would have taken literally hundreds of years on more conventional computer systems. We are now able to run virtual screens of chemical libraries against not a single protein target, but large panels of drug resistant mutant proteins of that viral target. The computing power of World Community Grid has also enabled us to increase the size of the libraries that we use in our virtual screens. Previously we were only able to screen libraries of a few thousand compounds. Today we typically screen combined libraries containing over 5 million compounds against each HIV protein target. This gives us a much better chance of finding good hit molecules.
A critical aspect of computational docking is how the interacting molecules are represented or modeled. AutoDock was the first docking code that allowed the model of the chemical compound to be flexible when docking to a protein target. Each rotatable bond in the compound added another dimension to the search space that had to be explored, and thus to the complexity of the computation. In those days, the target protein had to be treated as a rigid molecule, since the complexity of its flexibility would be too computationally difficult to simulate. Using the power of World Community Grid, we have been able to overcome this restriction, and have implemented different representations of protein flexibility, giving us more realistic models of the protein target. To this point FAAH volunteers have donated over 340,000 years of computing time to our research efforts.
Scientific accomplishments
By screening libraries of compounds against both wild type and numerous drug resistant mutant versions of HIV protease we have been able to characterize the range of the mutants that exist and cluster them into characteristic variants. This has given us a way to reduce the number of prototypic mutants of HIV PR that we need to use in our virtual screens, and has enabled us and others to focus our attention on the kinds of variations that the virus is capable of making to remain functional in the presence of drug therapies.
As the number of World Community Grid volunteers and the amount of computer power they donated increased through the years, we have been able to greatly increase the size and complexity of the types of experiments we could perform. We have now docked millions of small molecule compounds against several different regions of all of the enzymes that HIV produces—protease, reverse transcriptase, and integrase. HIV reverse transcriptase is the part of the viral machinery that makes new copies of its genetic material. It takes the HIV RNA from the original viral particle and makes a more stable DNA version of it (called cDNA, or complementary DNA). HIV integrase then processes the viral DNA, by clipping part of each end off of it, to generate more reactive “sticky ends.” Integrase then permanently attaches the sticky ends of the viral DNA into our human DNA, to create a permanently infected cell, which will then go on to produce new copies of the virus. Thus, World Community Grid enabled us to computationally evaluate millions of compounds against many different regions of all of the viral machinery—instead of only being able to evaluate thousands of compounds against one region of one type of enzyme. It will take us many years to analyze these mountains of data, select the most promising candidate compounds, and experimentally evaluate these candidates in “wet lab” experiments performed by our collaborators. But we have created new tools to help us process and analyze these data more efficiently, and we are developing additional approaches to help us harvest more useful and more accurate information from the results that volunteers like you have produced.
Most recently, we have turned our attention to the concept of synergistic inhibition of HIV enzyme function. In addition to targeting the active sites of these enzymes, we have also been able to search for new inhibitors that bind to “allosteric” sites on each of these viral nanomachines. Allosteric inhibitors have been discovered in both HIV integrase and reverse transcriptase. Most inhibitors disable the function of an enzyme by binding directly to the “active site” (the specific region of an enzyme where the chemical work occurs) and blocking its ability to function. The 9 FDA-approved HIV protease drugs are an example of these conventional types of inhibitors: they bind to and block the active site in the hollow tunnel in the center of protease, which prevents the viral multi-protein polypeptide from being able to bind within that tunnel and get cleaved. Allosteric inhibitors work in a very different way—they bind to a totally different site, regulate the conformational preferences and/or flexibility of the entire target protein, and thereby disable the active site. They bind far from the active site and project their influence over the rest of the enzyme. Allosteric inhibition is like putting a latch on the handles of a pair of scissors in order to prevent the blades from being able to open and close and cut things. Most importantly, different experiments with HIV, cancer, and malaria have shown that combinations of allosteric inhibitors and active site inhibitors can (a) generate combination therapies that are more effective against current superbugs and that (b) actually slow down the evolution of new drug-resistant mutants.
There are no known allosteric inhibitors of HIV PR. However we have postulated that there may exist sites for allosteric inhibition of HIV PR. We have run FAAH virtual screens against candidate sites, and have found that there are chemical compounds that can bind to these sites. These observations are bolstered by experimental x-ray diffraction experiments on PR in the presence of a chemical fragment library. Observed in these experiments are 3 sites that are distal to the active site of HIV PR where chemical fragments bind. We have now focused on finding better binding compounds at these sites using our FAAH virtual screens. Some of the hits from of these screens have been verified by x-ray crystallography and NMR spectroscopy. Further development and optimization of these hits is ongoing. We are also working closely with other HIVE Center investigators, Prof. Kvaskhelia at Ohio State University, and Prof. Arnold at Rutgers University on improving allosteric inhibition for HIV integrase and reverse transcriptase, respectively.
As stated above, we have only scratched the surface in terms of analyzing the enormous data sets that have been produced on World Community Grid. Typically we have only selected and looked at the very top candidates from a large virtual screen. Since the energy evaluation from docking is only a crude approximation of the true energy of binding, we, in fact could be selecting compounds that are not the best binders – so-called "false positives." If we suggest to our collaborators to synthesize or buy such compounds, it can be very costly in time and money. Thus we are starting several efforts to reduce the number of false positives that arise from our virtual screens. Some of these efforts involve data mining of all of the FAAH results, looking at the interaction patterns of all of the compounds that are docked. This data can give us some chemical signatures that allow us to filter out the false positives from the true binders. This is ongoing work, but initial results look very promising.
Refining results
Another approach to this problem that has been developed in collaboration with the Levy Lab at Temple University involves a separate computational step that evaluates not just the top-scoring compound from a virtual screen, but many of the top ranked compounds. This computation uses sophisticated molecular dynamics based estimates of the free energy of binding, which are too computationally difficult to perform during the docking, but which can discriminate between true and false positives subsequent to the docking. This approach, called BEDAM, is being implemented with a molecular simulation tool called Academic IMPACT which is running on Phase 2 of our FAAH project on World Community Grid.
In summary, World Community Grid volunteers have enabled an invaluable resource for our ongoing work in understanding the nature of drug resistance in HIV. As with most scientific projects, along with new results, new questions arise. Although we have yet to get a complete picture of the evolution of HIV drug resistance or create a new drug that defeats HIV resistance, we have developed new computational methodologies and have opened a number of avenues of research, that may lead to better approaches to treating and possibly even curing HIV infection.
A sincere thank you to all of the volunteers who donated their time to this project. And while the initial phase of the FightAIDS@Home project has come to an end, be sure to contribute to the second phase of the project.
Related Articles
- The FightAIDS@Home project adds new modeling tool
- Pioneering new techniques in the fight against HIV
- FightAIDS@Home Team Expands Techniques, Refines Phase 1 Results, and Collaborates on a New Study
- FightAIDS@Home Team Re-Opens Phase 1
- FightAIDS@Home Has a New Tool and a New Teammate
- FightAIDS@Home – Phase 2 Prepares for A New Stage