In this project update, the Mapping Cancer Markers team describes how they are analyzing 45 million of the most promising lung cancer data results, and how they have begun to disseminate their early findings.
The Mapping Cancer Markers (MCM) project continues to process work units for the Ovarian Cancer dataset. As we accumulate these results, we continue to analyze MCM results from the previous Lung Cancer dataset. Below, we discuss one direction in which we are pursuing the analysis.
Patterns of gene-family signatures in lung cancer
In cancer, and human biology in general, multiple biomarkers (genes, proteins, microRNAs, etc.) can have similar patterns of activity. This may be because the genes serve redundant roles, or because the genes (or other molecules) participate together in a group to serve a biological function. A cancer signature composed of a set of specific genes may appear different than another signature composed of different, specific genes, and yet perform equivalently because the genes in each are functionally related. With this problem in mind, post-doctorate fellow Anne-Christin Hauschild is leading a study of frequently-occurring patterns (or motifs) of genes present in high-performing lung cancer gene signatures.
(Click on the picture below to see an enlarged version.)
This project looked at the first phase results from the Lung Cancer MCM analysis, which was a systematic exploration of the entire space of potential fixed-length signatures. We began by selecting 45 million high-performing signatures derived from World-Community-Grid-computed MCM results. These are the signatures evaluated to carry the most information for lung cancer diagnosis.
Next, we divided all genes in the lung cancer dataset into 180 clusters (gene families), where genes in each family show similar activity in the lung cancer dataset. We then labelled those top signatures with the gene families into which the genes were assigned. This gave us a set of high-performing signatures expressed as gene families instead of genes. This allowed us to treat two different gene signatures as the same gene-family signature, as long as the corresponding genes in each signature are members of the same family.
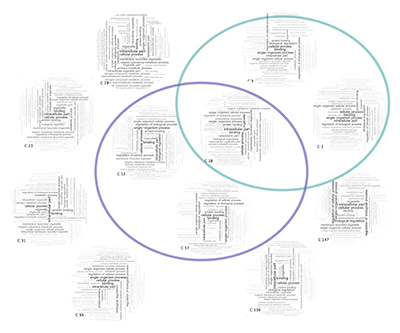
To help understand the gene-families themselves, we can visualize each one with word clouds that describe the functions of the genes they contain, or the biological pathways they represent. We draw this information from databases such as Gene Ontology, pathDIP, or other sources.
From there, we looked for patterns in these gene-family signatures: which families appear unusually frequently (or rarely) in high-performing signatures, or families that tend to appear multiple times in the same signature. We used Frequent-Itemset mining algorithm to discover specific patterns that occur unusually frequently in good signatures.
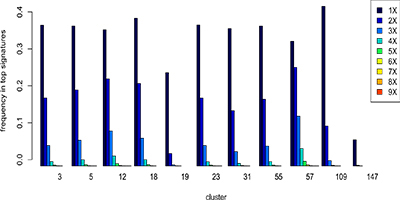
Illustration 2: Some gene families occur multiple times in a single signature with surprising frequency (high or low). Family 109 rarely appears multiple times. Family 12 appears surprisingly often in 9x multiples.
(Click on the picture below to see an enlarged version.)
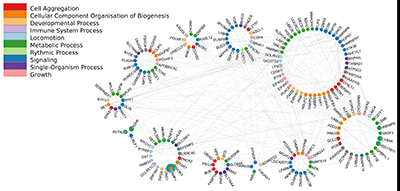
Using databases such as IID or pathDIP, we can take these patterns and examine the relationships between the gene-families they contain, so we can start to understand why certain combinations of such families carry so much information about lung cancer. We use NAViGaTOR to visualize and explore these complex sets of relationships.
(Click on the picture below to see an enlarged version.)
Early project results presented at Personalizing Cancer Medicine 2017
We presented the preliminary results of this project to Canadian and international cancer researchers this February, in a poster at the Personalizing Cancer Medicine Conference 2017 in Toronto, Ontario. We gained many insights and ideas from discussing this early work, and we continue developing them further.
Some of the additional, related results have been presented in other publications, including:
- Pinheiro, M., Drigo, S.A., Tonhosolo, R., Andrade, S.C.S., Marchi, F.A., Jurisica, I., Kowalski, L.P., Achatz, M.I., Rogatto, S.R., HABP2 p.G534E variant in patients with family history of thyroid and breast cancer, Oncotarget, In press.
- Citron, F., Armenia, J., Barzan, L., Franchin, G., Polesel, J., Talamini, R., Sulfaro, S., Croce, C.M., Klement, W., Pastrello, C., Jurisica, I., Vecchione, A., Belletti, B., Baldassarre, G., A microRNA signature identifies SP1 and TGFbeta pathways as potential mediators of local recurrences in head and neck squamous carcinomas, Clin Cancer Res, In press.
- Sokolina K, Kittanakom S, Snider J, Kotlyar M, Maurice P, Gandía J, Benleulmi-Chaachoua A, Tadagaki K, Wong V, Malty RH, Deineko V, Aoki H, Amin S, Riley L, Yao Z, Morató X, Otasek D, Kobayashi H, Menendez J, Auerbach D, Angers S, Pržulj N, Bouvier M, Babu M, Ciruela F, Jockers R, Jurisica I, and Stagljar I. Systematic protein-protein interaction mapping for clinically-relevant human GPCRs, Mol Sys Biol, In press.
- Yao Z, Darowski K, St-Denis N, Wong V, Offensperger F, Villedieu A, Amin S, Malty R, Aoki H, Guo H, Xu Y, Iorio C, Kotlyar M, Emili A, Jurisica I, Babu M, Neel B, Gingras AC, and Stagljar I, A global analysis of the protein phosphatase interactome, Mol Cell, in press.
- Petschnigg J, Kotlyar M, Blair L, Jurisica I, Stagljar I, and Ketteler R, Systematic identification of oncogenic EGFR interaction partners, J Mol Biol, in press.
- Rahmati, S., Abovsky, M., Pastrello, C., Jurisica, I. pathDIP: An annotated resource for known and predicted human gene-pathway associations and pathway enrichment analysis. Nucl Acids Res, 45(D1): D419-D426, 2016.
- Chehade, R., R. Pettapiece-Phillips, Salmena, L., Kotlyar, M., Jurisica, I., Narod, S. A., Akbari, M. R., Kotsopoulos, J. Reduced BRCA1 transcript levels in freshly isolated blood leukocytes from BRCA1 mutation carriers is mutation specific, Breast Cancer Res, 18(1): 87, 2016.
- Cierna, Z., Mego, M., Jurisica, I., Machalekova, K., Chovanec, M., Miskovska, V., Svetlovska, D., Hainova, K., Kajo, K., Mardiak, J., Babal, P. Fibrillin-1 (FBN-1) a new marker of germ cell neoplasia in situ, BMC Cancer, 16: 597, 2016.
Thank you to members
This work would not be possible without the participation of World Community Grid Members. Thank you for generously contributing CPU cycles, and for your interest in this and other World Community Grid projects.
Related Articles
- Project roadmap and first phase results from the Mapping Cancer Markers team
- Early-stage results from the Mapping Cancer Markers team
- Using one cancer to help defeat many: Mapping Cancer Markers makes progress
- One step closer to identifying lung cancer biomarkers
- Working to detect lung and ovarian cancers before they start
- Mapping Cancer Markers Now Examining Ovarian Cancer
- Mapping Cancer Markers Begins Analyzing Lung and Ovarian Cancer Data
- New Facility and Expanded Plans for the Mapping Cancer Markers Research Team
- Sarcoma Dataset Coming Soon to Mapping Cancer Markers Project
- Mapping Cancer Markers Researchers Analyzing Lung Cancer Data